what ethnic populations seem to have genetic immunity from aids
Abstract
Discernable genetic variation among people and populations has an important role in infectious disease epidemics, including that of acquired immune deficiency syndrome (AIDS). Genetic clan assay of several big AIDS cohorts implicate 14 AIDS brake genes, polymorphic variants in loci that regulate HIV-1 prison cell entry, acquired and innate immunity, and cytokine defenses to HIV-1. The influence and translational touch on of these genes on individual and population sensitivity to AIDS is considerable.
Chief
The scourge of AIDS, discovered iii decades ago, is the nearly devastating epidemic of our time. The cumulative expiry toll is approaching 25 million, and the World Health Organization estimates that 42 meg people today are infected with man immunodeficiency virus (HIV), a virus that, left untreated, kills 90% of its victims. Although considerable progress has occurred in the development of therapy for AIDS, the 16 available antiretroviral drugs are not always constructive, are frequently toxic and do not articulate virus from sequestered tissue reservoirs where HIV proviral genomes capable of rebound tin can linger for decades1,ii,3. The stiff drugs that slow the progress of AIDS are only now beingness distributed in the developing world, where they are virtually needed. HIV's replication efficiency, broad tissue dissemination and mutational plasticity in infected people has made vaccine development a considerable and still unsolved claiming4,5,6. For Africa, where 70% of the world'south HIV-positive individuals live and die, United nations Secretary General Kofi Annan recently named HIV/AIDS "one of the master obstacles of development itself"vii.
Although AIDS is not generally considered a genetic affliction, the considerable heterogeneity in the epidemic is at least partially determined by variants in genes that moderate virus replication and immunity5,8,ix. Not all people exposed to HIV-1 go infected, and those who do progress to AIDS-defining pathology at dissimilar time intervals. Rapid progressors succumb to AIDS in 1–five years whereas long-term 'nonprogressors' avert AIDS for upward to xx years. There are multiple AIDS-defining conditions, including Kaposi's sarcoma, lymphoma, Mycobacterium tuberculosis, Pneumocystis carinii pneumonia (PCP) and cytomegalovirus infections. Infected individuals have heterogeneity in the strength of their innate, humoral and cell-mediated immune responses likewise as differences in how they respond to antiretroviral treatment.
AIDS restriction genes
In the early 1980s, geneticists at the US National Cancer Institute's Laboratory of Genomic Diversity initiated a program to search the man genome for AIDS brake genes (ARGs; human genes with polymorphic variants that influence the outcome of HIV-1 exposure or infection10,11). We began past enlisting collaboration with epidemiologists who were establishing prospective AIDS cohort studies. These included American people in groups at risk for exposure to HIV-1: gay men, hemophiliacs exposed to clotting factor contaminated with HIV-1 earlier the introduction of HIV-ane antibody blood screening in 1984 and intravenous drug users who shared needles in areas of high HIV-ane incidence (Table 1). Clinical, virological and immunological information were assessed twice yearly for each individual through the course of his or her infection. We transformed viably frozen lymphocytes from study participants with Epstein-Barr virus to produce lymphoblastoid prison cell lines, a renewable source of Deoxyribonucleic acid for population-based DNA assessments. Almost 8,500 study participants comprise our present study population, 6,007 with lymphoblastoid cell lines and the balance with nonrenewable DNA specimens (Tabular array ane).
Nosotros selected candidate ARGs from the growing trunk of host factors nominated to be involved in HIV-i pathogenesis (Fig. 1). We resequenced selected human genes and adjacent regulatory regions to detect common single-nucleotide polymorphism (SNP) variants to genotype in the accomplice populations, and we examined case-control comparisons of SNP allele and genotype frequencies and genetic and linkage equilibrium tests to uncover genetic influences. For ARGs that influenced the charge per unit of AIDS onset in individuals infected with HIV-1, nosotros used Kaplan-Meyer survival curves and Cox proportionate adventure statistics to examine the effect of unlike genotypes on specific AIDS outcomes12,13,xiv.
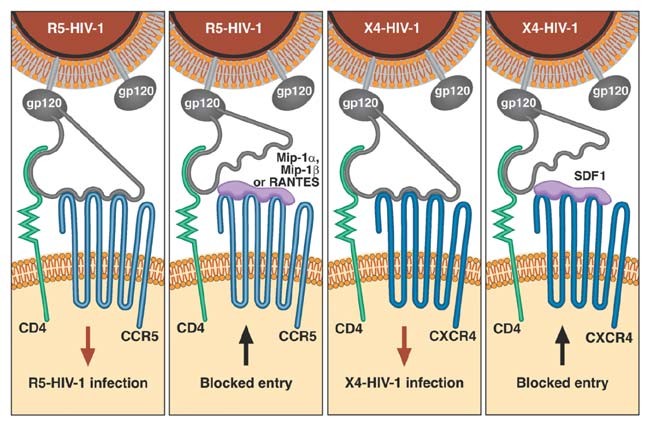
When HIV-1 infects a person, it seeks out tissue cell compartments where it can replicate, including lymph nodes, neural tissue, epithelium in gut or vagina, spleen, testes, kidneys and other organs. HIV'south principal factories are macrophages, monocytes and T-lymphocytes, all of which behave the CD4 cell surface poly peptide. HIV-1 enters diverse cells past co-opting two receptor proteins on the cell surface92,93. The CD4 molecule acts equally a docking station, which hooks the HIV surface envelope protein and leaves the virus hanging atop the cell. Then a large seven-transmembrane-spanning jail cell-surface receptor, CCR5, which floats effectually the fluid-like jail cell surface membrane, meanders into contact with the CD4-snagged HIV. The CCR5-HIV interaction causes the HIV gp41 poly peptide to change shape and penetrate the cell membrane; the HIV particle then fuses with the prison cell membrane and infects the cell. Infected individuals produce one to ten billion virus particles each day throughout the course of infection. The median time to CD4 cell collapse and onset of AIDS-defining pathologies is 10 years. In most infected people in Europe and The states (where HIV-one clade B is predominant), a mutational shift occurs in env (called R5 to X4 tropism), which alters coreceptor preference from CCR5 to CXCR4, commonly ancillary with CD4 depletion. The normal chemokine ligands specific for CCR5 (RANTES, MIP1α and Mip1β) and CXCR4 (SDF1) physically block CCR5 (R5) and CXCR4 (X4) HIV-ane infection, respectively, by roofing their entry coreceptorshalf dozen,23.
There were many rationales for identifying ARGs, just we predictable iii master applications. First, determining the effects of ARGs in individuals with AIDS would connect laboratory-identified host factors to AIDS pathogenesis explicitly, particularly when a plausible functional or physiological mechanism was found. Second, because the powerful antiretroviral therapy is neither universally effective nor a true cure, genetic retardation of AIDS progression could implicate new cellular targets for anti-AIDS therapy that complement the currently available antiretroviral compounds15. Tertiary, ongoing clinical trials for new vaccines and anti-AIDS drugs, similar longitudinal accomplice studies, evidence that these drugs elicit heterogeneous responses from people with AIDS. As some portion of this heterogeneity is attributable to the distribution of ARG genotypes in the report population, a quantitative ARG score, a 'genetic propensity index' (GPI) for individuals that could exist used to adapt (or subtract) the genetic 'dissonance' in clinical trials based on each participant's ARG genotype16, would be useful.
Since the first ARG variant, CCR5 Δ32, which effectively blocks HIV-i infection in homozygous people and slows AIDS progression in heterozygotes17,18,nineteen, was reported, our group and others accept documented 13 boosted ARGs through genetic association in these same cohort populations (Table two). Their genetic influences differ in several means, including fashion (dominant, codominant, recessive), stage at which they human action (infection, AIDS progression, the type of AIDS-defining pathogenic affliction) and the interval of HIV-1 progression during which the influence is apparent. Strategies for discovering ARGs and evidence for genetic association have been reviewedix,fourteen, as have the physiological mechanisms of ARG action8,sixteen, variation in ARG frequency and influence amidst indigenous groupstwenty,21 and replication in independent accomplice populationsviii,fourteen,22,23,24. ARGs are polygenic and multifactorial (i.e., they must collaborate with environmental factors, such as HIV-ane exposure, in order to be detected).
In this review, we explore the hallmarks of genetic association that indicated that these genes influence AIDS pathogenesis and the translational benefits of ARGs to therapy development. Nosotros talk over the quantitative influence of private and composite ARG genotypes in predicting AIDS outcomes for a person with HIV and for a study population. We estimate the influence that known ARGs have on the epidemic and compute a new information theory–based epidemiological parameter, the 'explained fraction' (EF, Box 2) to quantify the overall influence of multiple ARGs on the rate at which a population progresses to AIDS (ref. 25 and 1000.West.N. & S.J.O., unpublished data). The EF offers a quantitative yardstick that estimates the proportion of epidemiological variance in AIDS progression (or in whatsoever complex multifactorial disease) contributed by one or more ARGs in the written report population.
Validating ARGs in association studies
The ARGs in Table 2 were identified and validated past genetic association studies comparison alternative illness categories (e.yard., infected with HIV-one versus exposed to HIV-ane just not infected, or fast progressors versus slow progressors) for departure of allele, genotype or linkage equilibrium using the expectations of population genetic theory. Association analyses can exist misleading when statistical significance alone is the basis for factor identification, and there are likewise many examples of artifactual associations in the literature from studies that are underpowered statistically, are inappropriately structured with respect to ancestral population admixing or incorporate sampling errors26,27,28. To avoid mistaken associations, several authors have proposed benchmarks or stringent criteria for validation28,29.
For ARGs, there are five common aspects of a apparent genetic association: (i) High number of clinical cases, leading to low P values for significance after statistical correction for multiple tests. Ability calculations suggest that more than than 400 cases are adequate to detect a genotypic association with a relative risk (RR) ≥2.0. (ii) Independent replication of meaning associations in dissimilar populations, in different ethic groups, in different take a chance groups, in dissimilar cohorts and by unlike laboratories or investigators. If ARG allele frequencies differ among indigenous groups, the analysis should exist stratified to avoid population structure artifactsthirty,31. (3) Epidemiological influences that take high RR, loftier relative hazard (RH) and high attributable take a chance (AR, Box 1). Low risks mean the gene is either relatively unimportant or a statistical fluke. (four) Factor associations assessed alone, equally well as with a statistical adjustment for other previously associated gene types. This latter consideration tests epidemiological independence of multiple ARGs and uncovers epistatic interaction of genes with related physiological roles. (five) Plausible functional explanations that connect the operative SNP to a quantitative divergence in a gene production and chronicle the change to AIDS development. Although each of these five criteria were non fulfilled for every ARG in Table 2, almost ARGs met most of criteria. Their functional connections to AIDS progression are summarized in the side by side section.
Machinery of activity for ARGs
AIDS is primarily a disease of the immune system acquired by a virus evolved to infect, take over and debilitate the host'southward highly evolved innate and acquired immune defenses. Naturally occurring genetic variants in genes empirically connected to AIDS pathogenesis were discovered in the 2 decades of AIDS dissemination and study. The genetic epidemiological associations have been interpreted in the context of an implicated gene'southward function in development of AIDS, and in each instance a plausible functional mechanism has led to varying levels of empirical back up (Table 2).
The steps in establishing a chronic HIV-i infection leading to AIDS are illustrated in Figure 1. Genes encoding cellular molecules involved in the process were candidate ARGs. CCR5 Δ32 is a naturally occurring knockout deletion (32 bp) variant. Homozygotes resistant to R5-HIV-ane infection (the main infecting HIV-1 strain) lack the requisite HIV-one entry coreceptor CCR5 on their lymphoid cellsviii,17. Heterozygotes express less than half the wild-type levels of CCR5 receptors, slowing HIV-1 replication, spread and pathogenesis32,33. The CCR5 P1 allele, a composite haplotype comprising thirteen singled-out SNP alleles in the upstream promoter region of CCR5, confers recessive more rapid progression to AIDS34,35. Analysis of CCR5 promoter alleles found no quantitative differences in luciferase transcription, HIV-1 binding or HIV-ane infectivity, although the alleles influence CCR5 abundance on lymphoid cells36,37. Further, oligonucleotides that define the CCR5 P1 allele showed allele-specific recognition of nuclear transcription factors belonging to the cREL family, a result that suggests, but does not prove, that CCR5 promoter alleles could differentially react and answer to transcription factors in different cell types37.
The CCR2 V64I variant mediates filibuster in AIDS progression38,39 indirectly, as it causes no allele-specific quantitative differences in the corporeality of CCR2 produced, in the power of alternative allelic products to bind HIV-ane or in signal transduction with CCR2-specific ligandsxl,41. One report indicated that CCR2 I64 protein tin can preferentially dimerize with CXCR4 polypeptides (the HIV-1 receptor that replaces CCR5 as an entry receptor at later stages; Fig. 1) whereas the wild-type CCR2 peptides do not42. Though it is unconfirmed, this mechanism suggests that CCR2 V64I delays AIDS by limiting the transition from CCR5 to CXCR4 in infected individuals, a turning point in the collapse of the CD4–T lymphocyte cell population and a prelude to AIDS-defining disease23.
RANTES is a primary chemokine ligand for CCR5. levated circulating RANTES levels accept been detected in exposed individuals who avoid infection and also in people infected with HIV-1 who accept a delayed onset of AIDS43,44. I of the seven SNP variants found in CCL5 (encoding RANTES), In1.1C, is nested in an intronic regulatory sequence chemical element that has differential allele affinity to nuclear bounden proteins and whose transcription is downregulated past a factor of iv (ref. 45). A reduction in RANTES production in individuals carrying this allele leads to rapid AIDS progression, ostensibly by uncovering bachelor CCR5 that facilitates the replication and spread of HIV-1.
The CXCL12 3′A variant is a Thousand→A transition in the 3′ untranslated region of one of two alternatively spliced transcripts of CXCL12 (also called SDF1; ref. 46). Stromal-derived factor (SDF1) is the primary ligand for the belatedly-stage HIV-1 receptor CXCR4, and the 3′A variant is 37 bp downstream of two blocks whose sequences are 88% and 92% conserved, respectively, in mouse and human transcripts46,47. The relatively high sequence homology is a signal for selective constraints on mutational divergence of the region considering it is a putative recognition sequence for RNA- (or DNA-) binding regulatory factors. Although allele-specific cellular or virological differences have not been explicitly established, the delay in onset of AIDS observed in CXCL12 3′A homozygotes might issue from overproduction of SDF1 in certain tissue compartments, postponing the CCR5-CXCR4 transition. A synergistic protective consequence in individuals carrying CXCL12 3′A and CCR2 I64 would be consistent with this model46.
Quantitative functional allelic distinction has not been confirmed for ii ARGs: CXCR6 E3K, which is associated with late-state progression in individuals with AIDS and Pneumocystis carinii pneumonia, and haplotypes in the chromosome 17q gene cluster that contain iii SNP variants effectually the chemokine genes CCL2, CCL7 and CCL11 (refs. 48,49). MCP1 (encoded by CCL2) and MCP3 (encoded by CCL7) are requisite ligands for CCR2 and CCR3, respectively, and eotaxin (encoded by CCL11) is a ligand for CCR3, a chemokine receptor implicated in HIV-one infection of microglial cells in the cardinal nervous organization50,51. A few studies have tied MCP1, MCP3 and eotaxin to HIV-ane pathogenesis through interaction with HIV-1 directly or indirectly by stimulating jail cell division and migration of macrophages and dendritic cells52. Ane CCL2-CCL7-CCL11 haplotype, H7, which includes SNP variants in a 31-kb region, is associated with resistance to HIV-i infection, based on comparisons of highly exposed uninfected individuals with exposed infected individuals49.
Two ARGs, interleukin-x (IL10) and interferon-γ (IFNG), encode powerful cytokines that inhibit HIV-i replication. The IL10 5′A SNP variant involves a promoter region alteration that reduces IL10 transcription by a factor of ii–4 (ref. 53). IL10 5′A allele-specific synthetic oligonucleotides do not demark certain ETS-family unit transcription factors, which recognize the wild-blazon IL10 allele sequence. Heterozygosity and homozygosity with respect to IL10 5′A accelerates AIDS progression, probably owing to downregulation of the inhibitory IL10 cytokine53. IFNG includes a polymorphic promoter region, ane allele of which (−179T) is inducible by tumor necrosis factor (TNFα), whereas the wild-blazon allele (−179G) is not54. In African Americans, among whom the IFNG −179T allele has a 4% frequency (<1% in European Americans), individuals infected with HIV who are heterozygous with respect to IFNG −179T progress to AIDS more rapidly than IFNG −179G/Yard homozygotes, indicating that allele-specific inducibility confers a take a chance of rapid AIDS progression55.
The essential role of HLA, the human major histocompatibility complex, in detecting and presenting infectious amanuensis peptides to T-cells is well established. The HLA region includes 128 expressed genes, one-third of which participate in various armaments of human immunity56. HLA grade I (A,B and C) and 2 genes (DR, DQ and DP) have considerable allele variation between individuals and populations, which is probably a reflection of the natural selective influence by celebrated disease outbreaks that afflicted the ancestors of modern homo indigenous groups. The arable variation in HLA alleles provides a broad range for individual recognition of virus agents to which they had been exposed in the by, likewise as those to which they had not57,58.
HIV-1 infects various immune cells as the prelude to HIV-one replication, proliferation, spread and CD4-T lymphocyte damage. Because different HLA alleles specify cell-surface molecules with specific recognition sites for infectious agents59, differential HIV-1 peptide motif recognition can influence both the fourth dimension interval from infection to AIDS60 and the kinetics of HIV-1 adaptive escape from immune surveillance in an infected private61. For example, carriers of HLA-B * 35 experience rapid progression to AIDS and HLA-B * 35 homozygotes develop AIDS in one-half the median time that information technology takes for infected people without HLA-B * 35 to develop AIDS62. Molecules encoded past HLA-B * 35 comprise a serologically defined, closely related group with 2 singled-out peptide recognition specificities. HLA-B * 35-PY molecules recognize processed HIV peptides of 9 amino acids with proline in position 2 and tyrosine in position 9; HLA-B * 35-Px molecules nowadays peptides with proline in position 2 and various amino acids non including tyrosine in position 9 (ref. 63). Survival association analysis indicated that HLA-B * 35-mediated AIDS acceleration was entirely caused by HLA-B * 35-Px, as carriers of HLA-B * 35-PY adult AIDS at the same average charge per unit every bit individuals with no HLA-B * 35 alleles60. The HLA-B * 35-Px versus HLA-B * 35-PY distinction directly connected AIDS progression to HLA-B * 35-Px peptide recognition.
The mechanism by which HLA-B * 35-Px accelerates progression to AIDS does not seem to stem from a failure of the allele to stimulate cytotoxic T-cell recognition. Several amino acid residues in HIV gag proteins are HLA-B * 35-Px targets61, and there is an equivalent consecration of cytotoxic T lymphocytes (CTLs) in carriers of HLA-B * 35-Px as in carriers of HLA-B * 35-PY, indicating that the requisite HIV epitopes corresponding to HLA-B * 35-Px and HLA-B * 35-PY stimulate CTL induction64. A plausible hypothesis suggests that the HIV epitopes recognized by HLA-B * 35-Px deed equally an 'immunological decoy' that effectively induces CTLs that are ineffective in destroying cells infected with HIV-1, thereby monopolizing CTL defenses. In support of this possibility, carriers of HLA-B * 35-PY reduce HIV-one viral load far more than effectively than carriers of HLA-B * 35-Px, despite having an equivalent quantitative CTL response64.
HLA influence on HIV-1 sequence divergence was confirmed by examining the pattern of mutation frequency amid HIV-1 genomes in 473 Australians infected with HIV-1 (ref. 61). In that location was a dearth of sequence variants in regions of the HIV-ane genome with HLA recognition motifs that correspond to an infected person's HLA type just a mutational excess in regions with motifs recognized by HLA alleles that were associated with rapid progression, correlating HLA recognition of cognate HIV-1 motifs with AIDS survival. Two HLA alleles, HLA-B27 and HLA-B57 are consistently associated with a delayed onset of AIDSnine,65. These HLA genotypes seem to be more than effective in constraining HIV-1 mutational escape from immune clearance than other HLA alleles9,16.
The breadth of HLA allelic and immunological diversity is good for individuals with AIDS and for populations at adventure for AIDS. People who are homozygous with respect to one or more of their HLA class I genes (A, B or C) progress to AIDS much faster than individuals who are heterozygous with respect to HLA-A, HLA-B and HLA-C 62,66. The advantage of maximal HLA heterozygosity in individuals (or populations) infected with HIV-1 stems from maximizing the breadth of virus epitope recognition, driven to the farthermost by a swarm of HIV-ane mutational variants produced each day.
Some provocative HLA associations involving functionally relevant HLA allele groupings with HIV infection and progression take also been reported67,68. Certain HLA supertypes, grouped according to B-pocket contact residues of the antigenic peptide recognized by HLA class I alleles, prove pocket-sized associations with HIV transmission and with circulating set-point HIV concentration (viral load)68. Alleles of the HLA-Bw4 grouping, which comprises twoscore% of HLA-B alleles that share a seven–amino acrid motif, are associated with long-term nonprogressors, although the study reporting this was controversial65,67. HLA-Bw4 alleles serve as a ligand for sure natural killer cell receptors encoded by the killer immunoglobulin receptor (KIR) factor complex on chromosome 19 (ref. 69). The combination of HLA-Bw4 and KIR3DS1 has an epistatic protective influence on AIDS progression70. An interaction between the HLA-Bw4 ligand molecules and their respective 'activating' receptor 3DS1 on natural killer cells seems to facilitate clearance of HIV-ane-infected lymphocytes, slowing AIDS progression. In all, the immune response genes are fertile ground for discovery of important and powerful ARGs.
Implications of ARGs for AIDS therapy
The sixteen available anti-HIV-one drugs target two enzymes encoded in the HIV-1 genome: opposite transcriptase and proteasefifteen. Recent drug formulations that allow single daily doses have improved compliance and prognosis, but there is notwithstanding a pressing demand for amend, safer, cheaper and more than effective anti-AIDS drugs. Defining the early steps in HIV-1 infection and AIDS progression (Fig. ane) has invigorated the search for new approaches to AIDS handling71,72,73,74. Specific inhibitors of HIV-i entry into cells and integration in cellular chromosomes tin can block cellular factors that HIV-1 exploits in AIDS pathogenesis. After CD4, CCR5 and CXCR4 receptors were shown to catalyze HIV-1 infection (Fig. 1), the finding that mutational variants of receptors and their ligands stemmed HIV-1 infection and progression (Tabular array 2) raised hopes that synthetic compounds that blocked the process would exist suitable inhibitors of virus-cell interaction. Inhibitors of HIV-one entry accept been developed, which block CD4 bounden (PRO-542 and BMS0806), CCR5 binding (SCH-C, SCH-D, PRO-140, United kingdom-426, Britain-857), CXCR4 bounden (AMD-3100) and gp41-mediated membrane fusion (T-20, T-1249). All these drugs are in advanced stages of clinical trials15, and ane, T-xx, was approved for public utilize in March 2003. In improver, an inhibitor of HIV-one integrase (S-1360) has reached clinical setting based on efficacious results in vitro and in an animal model75. Although no human ARG that affects integration has been described to appointment, there is a compelling genetic evidence that the mouse locus Fv1 resists retroviral integration76.
New cell-based therapies accept the advantage of existence specific, existence nontoxic, in several cases, and targeting host jail cell factors whose genes do non mutate and evolve resistance rapidly in vivo similar HIV-1. Although HIV-1 tin evolve resistance to cell-based therapies, the genetic restrictions of variants in CCR5, CXCL12, CCL5 and other ARGs (Tabular array 2) suggest that inhibitors of HIV-1 entry may be effective in treating infected individuals. Finally, ARG variants like CCR5 Δ32 indicate that certain host cellular products are dispensable for normal evolution and amnestyviii, which bodes well for cell-based therapies that target their HIV-1 interest.
A genetic propensity index for composite ARG genotype
The estimation of clinical trials for new vaccines and anti-HIV drugs would benefit from a method to quantify the influence a particular ARG genotype might exert on the observed heterogeneity in participants in the trial. When considering the fourth dimension interval from HIV-i infection to the onset of AIDS, it is possible to measure the influence of single or blended ARGs on AIDS survival directly in our combined AIDS cohort populationsfourteen,16. To do this, we computed the RH of culling genotypes relative to the common 'wild-type' genotype (Table 3) based on the Cox proportionate hazards model of Kaplan-Meyer survival curves12,13. For multiple loci RH is multiplicative, and the combined RH estimates the GPI for an private (Tabular array three). To illustrate the principle, we present private RHs and blended GPIs for eleven contained (in both epidemiologic and functional context) ARG genotypes (Table 3) and plot the predicted AIDS survival distribution (Fig. 2). An capricious baseline of GPI = 1.0 is the composite genotype carrying the wild-type genotype at each ARG locus.
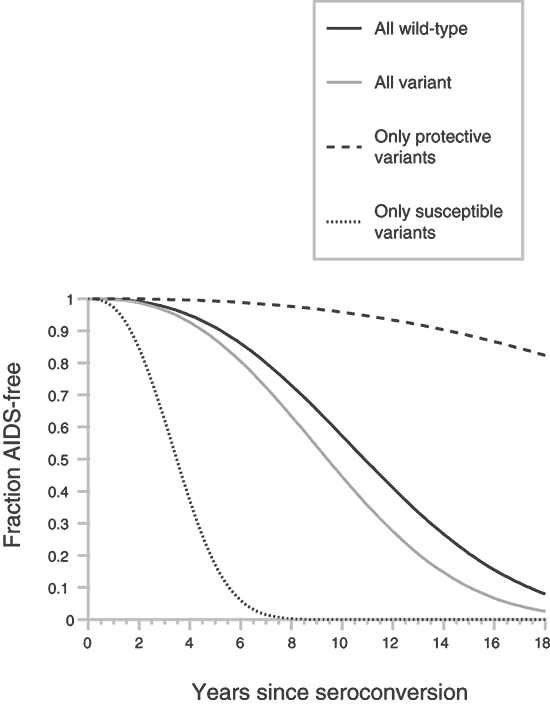
The Weibull distribution is used to model the survival distribution, with baseline shape and calibration parameters adamant from AIDS cohort information, taking RH for each group from the GPI for the given genotype combination.
The GPI for the ARG genotype that contains all the alternative 'non-wild-type' genotypes is i.46, non much different from baseline GPI, because the blended genotype containing all the variants includes five 'susceptible' and six 'resistant' genotypes, which offset each other in a survival distribution (Fig. 2). Composite GPIs for composites containing all xi susceptible genotypes (RH ≤1.0) or all eleven resistant genotypes (RH ≥i.0) define the extremes (GPI = 0.077 and 18.84, respectively) of predicted survival times (Table 3 and Fig. 2).
In a clinical trial, the GPI predicts the influence of the participants' genotypes on the trial outcome, in principle considering genotypes a confounding variable in the trial. The usefulness of such an application depends on the observed and theoretical variance of actual RH compared with GPI prediction for the study population. This variance affects sampling variance and the fraction of the overall epidemiological variance contributed by the genes we assess. Estimating this latter parameter is the adjacent topic.
Epidemiological influence of ARGs
The AR parameter can be used to quantify the influence of any epidemiological factor on disease incidence14,77,78. AR considers genetic mode (dominant, recessive, etc.) disease incidence and genotype frequency in disease cases (Box one). The AR for each protective ARG genotype on the charge per unit of AIDS progression (using Equation one) is given in Tabular array iv. The AR for genotypes that irksome AIDS progression ranged from two.iv% (for CCR2 I64) to half dozen.ix% (for CCR5 Δ32) for private genotypes. When all the genes that slow AIDS progression are considered equivalent, the cumulative AR for long-term survivors is 21.1%. This ways that 21.1% of individuals infected with HIV-1 who avoided AIDS for xi or more years did and then because they carried one or more than protective ARGs. Of the individuals who develop AIDS quickly, within 5.5 years of HIV-1 infection, the single ARG effects were modest (2.4–8.i%; Tabular array iv), but the cumulative AR for all the ARGs associated with rapid progression is considerable (twoscore.9%).
Although the overall AR values seem large, there are several limitations of the AR statistic78. Beginning, AR was designed for dichotomous factors, not multiple factors. This is a difficult complication considering offsetting susceptible and restricting ARGs in most people effectively cancel each other's influence. Second, AR values are not inherently additive; adding AR of multiple risk factors including genotypes volition often exceed 100%, distorting the actual influence. 3rd, AR is asymmetrical with respect to population distribution of a disease in because just the disease cases. AR measures whether a causal factor is necessary, simply does not consider whether it is sufficient, considering it does not consider the incidence of the factor in nondisease categories.
For these reasons, we used an epidemiologically symmetrical parameter, the EF, to assess the overall quantitative influence of multiple ARGs or other causal factors on AIDS survival distributions (Box 2). EF is a mutual information-based estimate of the influence of one or more causal factors (genetic or environmental) on disease incidence in the at-chance population; EF for each cistron is additive to a maximum of 100% (ref. 25 and Thou.Due west.North. & S.J.O., unpublished information). In Table 4, we present EFs for each individual ARG and compare the EF sum for all protective and susceptible genotype combinations with the ARs for the aforementioned genotypes. For context we too compute EFs in our cohorts (Table one) for the well-known nongenetic influence of age79,80.
The EF for each gene is modest, ranging from ≤0.1% (for CCR2 I64) to 2.4% (for HLA-B * 27). The EF for combined genotypes is computed for all combinations of 11 genotypes (73 different combinations). The EF for combined factors was seven.iv% for the ARG influence on rapid progressors to AIDS and 9.5% for the explanation of long-term survivors who avert AIDS for 11 years or more. The sum of the private ARG EF estimates is 7.2% for rapid progressors and 9.8% for long-term survivors, comparable to the combined ARG genotype EF estimates (G.N. & S.J.O., unpublished information). The EF for all ARGs plus historic period is 9.nine% for rapid progression and 12.vii% for slow progression (Table 4).
The modest EF has of import implications for the application of ARG genotype to prognosis of individuals with AIDS and to clinical trials of new therapeutics and AIDS vaccines. Because more than 90% of the epidemiological variation is not explained by the sum of ARG genotype, any weighting of study participants would have a large unexplained variance; the unexplained variance is ten times greater than the variance explained by the genes. This ways that predictions based on ARGs in accomplice studies or vaccine trials today would be imprecise and statistically noisy. This indicate is illustrated in Effigy 3, which shows the fourth dimension to AIDS predicted by GPI of 600 individuals plotted against the bodily time to AIDS for the same individuals or for dissimilar cohorts. Both regressions show a modest correlation (Rii = 0.14 for each; Fig. iii), which, though statistically significant, would hardly be useful in a clinical trial setting. As such, there is much left to explain in the varying influences on AIDS progression.
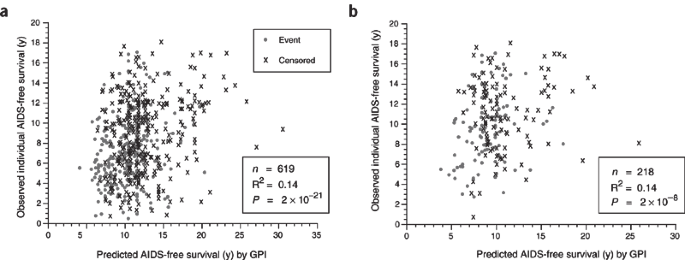
Actual AIDS-free survival91 versus the mean of the Weibull distribution for the subjects GPI and scale parameter, calculated by accelerated failure fourth dimension regression (SAS PROC LIFEREG). Subjects are European Americans in combined AIDS cohorts (Table 1). (a) Actual survival of all subjects versus predicted survival from GPI calculated from accelerated failure time regression on all subjects. (b) Survival of subjects in the Multicenter Hemophilia Cohort Report and San Francisco City Clinic Men'southward Study cohorts versus predicted survival from GPI calculated for subjects in Multicenter AIDS Cohort Study and Alive Study cohorts (Table ane).
These computations practice, notwithstanding, offer some optimistic conclusions near AIDS and gene discovery. Starting time, genes with small-scale RRs and EFs that have measurable quantitative effects on gene expression tin can be validated with high statistical confidence and replicated in independent cohort designs. Second, although the identified ARGs explain only a small-scale fraction of the variance, their EF (seven.3–9.4%; Table four) is comparable to that of smoking in lung cancer (nine.7–11.five%; Box ii). Tertiary, nosotros applied a quantitative yardstick to measuring the additive influence of multiple genes for a complex, multifactorial disease, a useful measure that tells u.s. how much more epidemiologic variation needs to be explained. Finally, the identified ARGs were based on SNP variation association in candidate genes, already continued to AIDS past the basic AIDS enquiry community. As less than 20% of the ∼25,000 human genes have names or suspected function, many unknown ARGs may exist.
Conclusions
The AIDS epidemic is a defining medical and public health result of our generation. Information technology has galvanized bones research in virology, vaccine evolution, antiviral therapy and health educational activity. In this review, we attempted to highlight important points in identifying human being genes that influence complex and multifactorial diseases, with the goal of applying these discoveries to further defining the regulatory components of an infected individual's cell physiology in AIDS progression and pathogenesis. The mechanism of AIDS restriction past natural genetic variants has encouraged the evolution of a new grade of anti-AIDS drugs, which block HIV-i binding to requisite receptors and membrane fusion (Fig. 1). Finally, we explore the potential application of predicting AIDS progression for a given genotype combination to inform clinical trials for new drugs and vaccines. As with genes associated with susceptibility to any complex, chronic disease, the hope is to inform diagnostics, prevention, vaccine and treatment development. For each awarding, the ARGs fulfill some only not all expectations, emphasizing the need for further factor identification and interpretation.
We examine two new epidemiology parameters. GPI is a quantitative score based on empirically observed epidemiological survival that predicts the RH and survival of different ARG combination genotypes (Tabular array 3 and Fig. 2). This weighting is an early footstep toward predicting AIDS outcome among infected individuals past considering the survival influence of the genotypes they carry. EF is an data theory–based parameter designed to estimate the portion of epidemiological variance in an epidemic that is due to a particular ARG genotype or composite multilocus genotype. A modest percentage (∼ix%) of the variation in AIDS progression kinetics are explained past the sum of ARGs at present known, barrel that fraction is comparable to the same guess of smoking influence on lung cancer (9.7–11.5%), a well-known epidemiological gene.
The nexus of epidemiological patient cohorts and population genetic–based association approaches has shown that gene-based epidemiological assay is a powerful tool for implicating multiple associated loci in circuitous illness. The identification and validation of the ARGs in Table two offer hope that additional genes not nonetheless known will exist discovered non only amid known candidate genes, simply likewise by SNP haplotype-based clan in high-density genome scans of clinically well-described epidemiological cohorts81. Nosotros estimate that more than 90% of the genetic and nongenetic influence on AIDS progression in people infected with HIV-i is undiscovered. Yet, the validation and interpretation of the fourteen known ARGs lends credence to the hereafter search by genome-broad association scans of available cohort populations. The consequences of farther investigation will be valuable if their translational benefits include improved diagnostics and treatment for AIDS.
References
-
Palella, F.J. Jr. et al. Declining morbidity and mortality among patients with advanced human being immunodeficiency virus infection. HIV outpatient Study Investigators. Northward. Engl. J. Med. 338, 853–860 (1998).
-
Mocroft, A. et al. Changing patterns of mortality across Europe in patients infected with HIV-i. EuroSIDA Study Group. Lancet 352, 1725–1730 (1998).
-
Finzi, D. et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on constructive combination therapy. Nat. Med. 5, 512–517 (1999).
-
Cohen, J. Shots in the Dark. The wayward search for an AIDS vaccine. (Westward. Norton, New York, 2001).
-
Fauci, A.S. HIV and AIDS: twenty years of science. Nat. Med. 9, 839–843 (2003).
-
Weiss, R.A. HIV and AIDS: looking ahead. Nat. Med. 9, 887–891 (2003).
-
Annan, K. Forward. in AIDS in Africa (eds. Essex, Thousand., Mboup, Due south., Kanki, P.J., Marlink, R. & Tlou, S.D.) 2nd edn. (Kluwer Academic, New York, 2002).
-
O'Brien, Due south.J. & Moore, J. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177, 99–111 (2000).
-
Carrington, Yard. & O'Brien, S.J. The influence of HLA genotype on AIDS. Ann. Rev. Med. 54, 535–551 (2003).
-
O'Brien, Southward.J. & Dean, G. In search of AIDS-resistance genes. Sci. Am. 277, 44–51 (1997).
-
O'Brien, Southward.J. AIDS: A function for host genes. Hosp. Pract. 33, 53–79 (1998).
-
Cox, D.R. & Oakes, D. Analysis of Survival Data, London (Chapman and Hall, 1984).
-
Allison, P.D. Survival analyses using the SAS system: A applied guide. 155–197 (SAS Institute, Cary, N Carolina, 1995).
-
O'Brien, Southward.J., Nelson, G.West., Winkler, C.A. & Smith, M.West. Polygenic and multifactorial disease gene association in human being: Lessons from AIDS. Annu. Rev. Genet. 34, 563–591 (2000).
-
Gulick, R.M. New antiretroviral drugs. Clin. Microbiol. Infect. ix, 186–193 (2003).
-
Carrington, M., Nelson, Grand. & O'Brien, S.J. Considering genetic profiles in functional studies of immune responsiveness to HIV-1. Immunol. Lett. 79, 131–140 (2001).
-
Dean, Chiliad. et al. Genetic restriction of HIV-i infection and progression to AIDS by a deletion allele of the CKR5 structural factor. Science 273, 1856–1862 (1996).
-
Liu, R. et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-ane infection. Cell 86, 367–377 (1996).
-
Samson, M. et al. Molecular cloning and functional expression of a new homo CC chemokine receptor gene. Biochemistry 35, 3362–3367 (1996).
-
Winkler, C.A. & O'Brien, S.J. AIDS restriction genes in human ethnic groups: An cess. in AIDS in Africa (eds. Essex, M., Mboup, S., Kanki, P.J., Marlink, R. & Tlou, Southward.D.) 2nd edn. (Kluwer Bookish, New York, 2002).
-
Winkler, C., Ping, A. & O'Brien, S.J. Patterns of ethnic diversity amidst the genes that influence AIDS. Hum. Mol. Genet. thirteen, R9–R19 (2004).
-
D'Souza, Grand.P. & Harden, V.A. Chemokines and HIV-i second receptors - confluence of two fields generates optimism in AIDS research. Nat. Med. 2, 1293–1300 (1996).
-
Berger, E.A., Murphy, P.M. & Farber, J.M. Chemokine receptors as HIV-1 coreceptors: Roles in Viral Entry, Tropism, and Disease. Annu. Rev. Immunol. 17, 657–700 (1999).
-
McNicholl, J.1000., Smith, D.K., Qari, S.H. & Hodge, T. Host genes and HIV: The role of the chemokine receptor gene CCR5 and its allele (?32 CCR5) Emerging. Infect. Dis. 3, 261–271 (1997).
-
Schemper, M. Predictive accuracy and explained variation. Stat. Med. 22, 2299–2308 (2003).
-
Risch, Northward. & Merikangas, K. The future of genetic studies of complex man diseases. Science 273, 1517–1517 (1996).
-
Pritchard, J.1000., Stephens, M., Rosenberg, Due north.A. & Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 67, 170–181 (2000).
-
Glazier, A.M., Nadeau, J.H. & Aitman, T.J. Finding genes that underlie complex traits. Science 298, 2345–2349 (2003).
-
Anonymous. Freely associating. Nat. Genet. 22, ane–2 (1999).
-
Pritchard, J.Yard. & Donnelly, P. Case-control studies of association in structured or admixed populations. Theor. Popul. Biol. sixty, 227–237 (2001).
-
Falush, D., Stephens, D. & Pritchard, J.K. Inferences of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
-
Wu, L. et al. CCR5 levels and expression, pattern correlate with infectability past macrophage-tropic HIV-1, in vitro. J. Exp. Med. 185, 1681–1691 (1997).
-
Benkirane, M., Jin, D.Y., Chun, R.F., Koup, R.A. & Jeang, K-T. Machinery of transdominant inhibition of CCR5-mediated HIV-one infection by CCR5 Delta 32. J. Biol. Chem. 272, 30603–30606 (1997).
-
Martin, 1000.P. et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282, 1907–1911 (1998).
-
Carrington, Grand., Dean, Yard., Martin, K.P. & O'Brien, S.J. Genetics of HIV-ane infection: Chemokine receptor CCR5 polymorphism and its consequences. Hum. Mol. Genet. eight, 1939–1945 (1999).
-
Salkowitz, J.R. et al. CCR5 promoter polymorphism determines macrophage CCR5 density and magnitude of HIV-ane propagation in vitro. Clin. Immunol. 108, 234–240 (2003).
-
Bream, J.H. et al. CCR5 promoter alleles distinguished past specific Deoxyribonucleic acid bounden factors. Science 284, 223a (1999).
-
Smith, Chiliad.W. et al. Contrasting genetic influence of CCR2 and CCR5 receptor gene variants on HIV-1 infection and illness progression. Science 277, 959–965 (1997).
-
Anzala, A.O. et al. CCR2-64I allele and genotype clan with delayed AIDS progression in African women. Lancet 351, 1632–1633 (1998).
-
Lee, B. et al. Influence of the CCR2-V64I polymorphism on human immunodeficiency virus type 1 coreceptor action and on chemokine receptor function of CCR2b, CCR3, CCR5, and CXCR4. J. Virol. 72, 7450–7458 (1998).
-
Mariani, R. et al. CCR2-64I polymorphism is not associated with altered CCR5 expression or coreceptor function. J. Virol. 73, 2450–2459 (1999).
-
Mellado, One thousand., Rodriguez-Frade, J.M., Vila-Coro, A.J., de Ana, A.M. & Martinez, A.C. Chemokine control of HIV-i infection. Nature 400, 723–724 (1999).
-
Zagury, D. et al. C-C chemokines, pivotal in protection against HIV type one infection. Proc. Natl. Acad. Sci. The states 95, 3857–3861 (1998).
-
Gallo, R.C., Garzino-Demo, A. & DeVico, A.L. HIV infection and pathogenesis: what about chemokines? J. Clin. Immunol. 19, 293–299 (1999).
-
An, P. et al. Modulation of HIV infection and illness progression by interacting RANTES variants. Proc. Natl. Acad. Sci USA 99, 10002–10007 (2002).
-
Winkler, C. et al. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine cistron variant. Science 279, 389–393 (1998).
-
Bleul, C.C. et al. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature 382, 829–833 (1996).
-
Duggal, P. et al. Genetic influence of CXCR6 chemokine receptor alleles on PCP-mediated AIDS progression amongst African-Americans. Genes Immun. 4, 245–250 (2003).
-
Modi, W.S. et al. Directional selection in the MCP-i MCP-3 Eotaxin gene cluster in influences HIV-1 manual. AIDS 17, 2357–2365 (2003).
-
Bagiolini, 1000. et al. IL-8 and related chemotactic cytokines-CXC and CC chemokines. Adv. Immunol. 55, 97–179 (1994).
-
He, J.50. et al. CCR3 and CCR5 are co-receptors for HIV-ane infection of microglia. Nature 385, 645–649 (1997).
-
Sozzani, S. et al. Differential regulation of chemokine receptors during dendritic cell maturation: a model for their trafficking properties. J. Immunol. 161, 1083–1086 (1998).
-
Shin, H.D. et al. Genetic restriction of HIV-1 infection and AIDS progression by promoter alleles of interleukin x. Proc. Natl. Acad. Sci. USA. 97, 14467–14472 (2000).
-
Bream, J.H., An, P., Zhang, X., Winkler, C.A. & Young, H.A. A unmarried nucleotide polymorphism in the proximal IFN-gamma promoter alters control of gene transcription. Genes Immun. iii, 165–169 (2002).
-
An, P. et al. A TNF-inducible promoter variant of interferon-gamma accelerates CD4 T cell depletion in HIV-i infected individuals. J. Infect. Dis. 188, 228–231 (2003).
-
Aguado, B. et al. Consummate sequence and factor map of a homo major histocompatibility circuitous. The MHC sequencing consortium. Nature 401, 921–923 (1999).
-
Parham, P. & Ohta, T. Population biology of antigen presentation by MHC class I molecules. Science 272, 67–74 (1996).
-
Hughes, A.L. & Yeager, M. Natural selection at major histocompatibility circuitous loci of vertebrates. Annu. Rev. Genet. 32, 415–435 (1998).
-
Hairdresser, 50.D. et al. Overlap in the repertoires of peptides bound in-vivo by a group of related Class-I HLA-B allotypes. Curr. Biol. 5, 179–190 (1995).
-
Gao, X. et al. Effect of a unmarried amino acid alter in MHC grade I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344, 1668–1675 (2001).
-
Moore, C.B. et al. Evidence of HIV-i accommodation to HLA-restricted immune response at a population level. Science 296, 1439–1443 (2003).
-
Carrington, M. et al. HLA and HIV-ane: Heterozygote advantage and B * 35-Cw * 04 disadvantage. Scientific discipline 283, 1748–1752 (1999).
-
Steinle, A. et al. Motif of HLA-B*3503 peptide ligands. Immunogenetics 43, 105–107 (1996).
-
Jin, X. et al. Human being immunodeficiency virus Type one (HIV-1)-specific CD8+-T-Prison cell responses for groups of HIV-1-infected individuals with unlike HLA-B*35 genotypes. J. Virol. 76, 12603–12610 (2002).
-
O'Brien, South.J., Gao, X. & Carrington, M. HLA and AIDS: A cautionary tale. Trends Mol. Med. seven, 379–381 (2002).
-
Tang, J.M. et al. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus blazon I infection. AIDS Res. Hum. Retroviruses fifteen, 317–324 (1999).
-
Flores-Villanueva, P.O. et al. Command of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. USA 98, 5140–5145 (2001).
-
Trachtenberg, Eastward. et al. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. nine, 928–935 (2003).
-
Lanier, L.L. NK cell receptors. Annu. Rev. Immunol. 16, 359–393 (1998).
-
Martin, M.P. et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31, 429–434 (2002).
-
Cairns, J.Due south. & D'Souza, Grand.P. Chemokines and HIV-ane second receptors: The therapeutic connexion. Nat. Med. 4, 563–568 (1998).
-
O'Brien, S.J. A new approach to therapy. HIV Newsline 4, 3–6 (1998).
-
LaBranche, C.C. et al. HIV fusion and its inhibition. Antiviral Res. 50, 95–115 (2001).
-
Stevenson, M. New targets for inhibitors of HIV-i replication. Nat. Rev. Mol. Cell Biol. 1, 40–49 (2000).
-
Hazuda, D.J. et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287, 646–650 (2000).
-
Best, Due south., Le Tissier, P., Towers, G. & Stoye, J.P. Positional cloning of the mouse retrovirus restriction gene Fv-1. Nature 382, 826–829 (1996).
-
Levin, M.L. The occurrence of lung cancer in human being. Acta Unio Int. Contra Cancrum 9, 531–541 (1953).
-
Benichou, J. Attributable Risk. in Encyclopedia of Statistics. (eds. Armitage, P. & Colton, T.) (Wiley, New York, 1998).
-
Goedert, J.J. et al. A prospective study of human immunodeficiency virus blazon 1 infection and the development of AIDS in subjects with hemophilia. N. Engl. J. Med. 321, 1141–1148 (1989).
-
Darby, S.C., Doll, R., Thakrar, B., Rizza, C.R. & Cox, D.R. Time from infection with HIV to onset of AIDS in patients with haemophilia in the United kingdom of great britain and northern ireland. Stat. Med. 6, 681–689 (1990).
-
The International Hap Map Project. Nature 426, 789–796 (2003).
-
Phair, J. et al. Caused-Allowed-Deficiency-Syndrome occurring within 5 years of infection with Human-Immunodeficiency-Virus Type-1 - the Multicenter AIDS Cohort Written report. J. Acquir. Allowed Defic. Syndr. 5, 490–496 (1992).
-
Detels, R. et al. For the Multicenter AIDS Accomplice Study: Resistance to HIV-ane infection. J. Acquir. Immune Defic. Syndr. vii, 1263–1269 (1994).
-
Kaslow, R.A. et al. The Multicenter AIDS Cohort Written report - Rationale, organization, and selected characteristics of the participants. Am. J. Epidemiol. 26, 310–318 (1987).
-
Buchbinder, S.P., Katz, K.H., Hessol, N.A., O'Malley, P.Grand. & Holmberg, South.D. Long-term HIV-1 infection without immunologic progression. AIDS eight, 1123–1128 (1994).
-
Goedert, J.J. et al. Decreased helper lymphocytes-T in homosexual men. 1. Sexual contact in high-incidence areas for the Caused Immunodeficiency Syndrome. Am. J. Epidemiol. 121, 629–636 (1985).
-
Hilgartner, M.W. et al. Hemophilia growth and development study - design, methods, and entry data. Am. J. Ped. Hematol. Oncol. 15, 208–218 (1993).
-
Lederman, Thousand.M. et al. Human immunodeficiency virus (HIV) type 1 infection status and in vitro susceptibility to HIV infection among high-gamble HIV-1-seronegative hemophiliacs. J. Infect. Dis. 172, 228–231 (1995).
-
Vlahov, D. et al. Association of drug injection patterns with antibody to human immunodeficiency virus type 1 amongst intravenous durg users in Baltimore, Maryland. Am. J. Epidemiol. 132, 847–856 (1990).
-
Dean, K. et al. Reduced hazard of AIDS lymphoma in individuals heterozygous for the CCR5-?32 mutation. Cancer Res. 59, 3561–3564 (1999).
-
US Centers for Disease Control. Morb. Mortal. Wkly. Rep. 36 Suppl 1 (1987).
-
Goldsmith, Thou.A. & Doms, R.W. HIV entry: are all receptors created equal? Nat. Immunol. three, 709–710 (2002).
-
Littman, D.R. Chemokine receptors: keys to AIDS pathogenesis? Cell 93, 677–680 (1998).
-
Kullback, South. Information Theory and Statistics (Dover, Mineola, New York, 1968).
-
Nilsson, S., Carstensen, J.M. & Pershagen, One thousand. Bloodshed amidst male and female smokers in Sweden: a 33 year follow upward. J. Epidemiol. Customs Health 55, 825–830 (2001).
Author data
Affiliations
Corresponding author
Rights and permissions
Most this article
Cite this commodity
O'Brien, S., Nelson, G. Human genes that limit AIDS. Nat Genet 36, 565–574 (2004). https://doi.org/10.1038/ng1369
-
Received:
-
Accustomed:
-
Published:
-
Issue Appointment:
-
DOI : https://doi.org/10.1038/ng1369
Further reading
Source: https://www.nature.com/articles/ng1369
0 Response to "what ethnic populations seem to have genetic immunity from aids"
Post a Comment